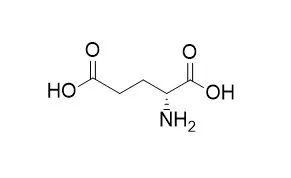
| Structure Identification: |
| Journal of Chromatography B, 2011, 879(29):3196-3202. | | Simultaneous determination of d-aspartic acid and d-glutamic acid in rat tissues and physiological fluids using a multi-loop two-dimensional HPLC procedure.[Reference: WebLink] | For a metabolomics study focusing on the analysis of aspartic and glutamic acid enantiomers, a fully automated two-dimensional HPLC system employing a microbore-ODS column and a narrowbore-enantioselective column was developed.
METHODS AND RESULTS:
By using this system, a detailed distribution of d-Asp and d-Glu besides l-Asp and l-Glu in mammals was elucidated. For the total analysis concept, the amino acids were first pre-column derivatized with 4-fluoro-7-nitro-2,1,3-benzoxadiazole (NBD-F) to be sensitively and fluorometrically detected. For the non-stereoselective separation of the analytes in the first dimension a monolithic ODS column (750 mm × 0.53 mm i.d.) was adopted, and a self-packed narrowbore-Pirkle type enantioselective column (Sumichiral OA-2500S, 250 mm × 1.5 mm i.d.) was selected for the second dimension. In the rat plasma, RSD values for intra-day and inter-day precision were less than 6.8%, and the accuracy ranged between 96.1% and 105.8%. The values of LOQ of d-Asp and d-Glu were 5 fmol/injection (0.625 nmol/g tissue).
CONCLUSIONS:
The present method was successfully applied to the simultaneous determination of free aspartic acid and glutamic acid enantiomers in 7 brain areas, 11 peripheral tissues, plasma and urine of Wistar rats. Biologically significant d-Asp values were found in various tissue samples whereas for d-Glu(D-Glutamic acid) the values were very low possibly indicating less significance. | | Journal of Bacteriology, 1993, 175(1):111-116. | | The Escherichia coli mutant requiring D-glutamic acid is the result of mutations in two distinct genetic loci.[Reference: WebLink] | D-Glutamic acid is an essential component of bacterial cell wall peptidoglycan in both gram-positive and gram-negative bacteria. Very little is known concerning the genetics and biochemistry of D-glutamate production in most bacteria, including Escherichia coli.
METHODS AND RESULTS:
Evidence is presented in this report for the roles of two distinct genes in E. coli WM335, a strain which is auxotrophic for D-glutamate. The first gene, which restores D-glutamate independence in WM335, was mapped, cloned, and sequenced. This gene, designated dga, is a previously reported open reading frame, located at 89.8 min on the E. coli map. The second gene, gltS, is located at 82 min. gltS encodes a protein that is involved in the transport of D- and L-glutamic acid into E. coli, and the gltS gene of WM335 was found to contain two missense mutations.
CONCLUSIONS:
To construct D-glutamate auxotrophs, it is necessary to transfer sequentially the mutated gltS locus, and then the mutated dga locus into the recipient. The sequences of the mutant forms of both dga and gltS are also presented. |
|



 Cell. 2018 Jan 11;172(1-2):249-261.e12. doi: 10.1016/j.cell.2017.12.019.IF=36.216(2019)
Cell. 2018 Jan 11;172(1-2):249-261.e12. doi: 10.1016/j.cell.2017.12.019.IF=36.216(2019) Cell Metab. 2020 Mar 3;31(3):534-548.e5. doi: 10.1016/j.cmet.2020.01.002.IF=22.415(2019)
Cell Metab. 2020 Mar 3;31(3):534-548.e5. doi: 10.1016/j.cmet.2020.01.002.IF=22.415(2019) Mol Cell. 2017 Nov 16;68(4):673-685.e6. doi: 10.1016/j.molcel.2017.10.022.IF=14.548(2019)
Mol Cell. 2017 Nov 16;68(4):673-685.e6. doi: 10.1016/j.molcel.2017.10.022.IF=14.548(2019)