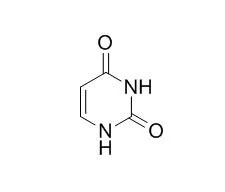
| Structure Identification: |
| J Biol Chem. 2014 Aug 8;289(32):22008-18. | | Excision of uracil from transcribed DNA negatively affects gene expression.[Pubmed: 24951587] | Uracil is an unavoidable aberrant base in DNA, the repair of which takes place by a highly efficient base excision repair mechanism. The removal of Uracil from the genome requires a succession of intermediate products, including an abasic site and a single strand break, before the original DNA structure can be reconstituted. These repair intermediates are harmful for DNA replication and also interfere with transcription under cell-free conditions. However, their relevance for cellular transcription has not been proved.
METHODS AND RESULTS:
Here we investigated the influence of Uracil incorporated into a reporter vector on gene expression in human cells. The expression constructs contained a single Uracil opposite an adenine (to mimic dUTP misincorporation during DNA synthesis) or a guanine (imitating a product of spontaneous cytosine deamination). We found no evidence for a direct transcription arrest by Uracil in either of the two settings because the vectors containing the base modification exhibited unaltered levels of enhanced GFP reporter gene expression at early times after delivery to cells. However, the gene expression showed a progressive decline during subsequent hours.
CONCLUSIONS:
In the case of U:A pairs, this effect was retarded significantly by knockdown of UNG1/2 but not by knockdown of SMUG1 or thymine-DNA glycosylase Uracil-DNA glycosylases, proving that it is base excision by UNG1/2 that perturbs transcription of the affected gene. By contrast, the decline of expression of the U:G constructs was not influenced by either UNG1/2, SMUG1, or thymine-DNA glycosylase knockdown, strongly suggesting that there are substantial mechanistic or kinetic differences between the processing of U:A and U:G lesions in cells. | | Phys Chem Chem Phys. 2014 Sep 7;16(33):17835-44. | | Anionic derivatives of uracil: fragmentation and reactivity.[Pubmed: 25036757] | Uracil is an essential biomolecule for terrestrial life, yet its prebiotic formation mechanisms have proven elusive for decades. Meteorites have been shown to contain Uracil and the interstellar abundance of aromatic species and nitrogen-containing molecules is well established, providing support for Uracil's presence in the interstellar medium (ISM). The ion chemistry of Uracil may provide clues to its prebiotic synthesis and role in the origin of life. The fragmentation of biomolecules provides valuable insights into their formation. Previous research focused primarily on the fragmentation and reactivity of cations derived from Uracil.
METHODS AND RESULTS:
In this study, we explore deprotonated Uracil-5-carboxylic acid and its anionic fragments to elucidate novel reagents of Uracil formation and to characterize the reactivity of Uracil's anionic derivatives. The structures of these fragments are identified through theoretical calculations, further fragmentation, experimental acidity bracketing, and reactivity with several detected and potential interstellar species (SO2, OCS, CS2, NO, N2O, CO, NH3, O2, and C2H4). Fragmentation is achieved through collision induced dissociation (CID) in a commercial ion trap mass spectrometer, and all reaction rate constants are measured using a modification of this instrument.
CONCLUSIONS:
Experimental data are supported by theoretical calculations at the B3LYP/6-311++G(d,p) level of theory. Lastly, the astrochemical implications of the observed fragmentation and reaction processes are discussed. |
|



 Cell. 2018 Jan 11;172(1-2):249-261.e12. doi: 10.1016/j.cell.2017.12.019.IF=36.216(2019)
Cell. 2018 Jan 11;172(1-2):249-261.e12. doi: 10.1016/j.cell.2017.12.019.IF=36.216(2019) Cell Metab. 2020 Mar 3;31(3):534-548.e5. doi: 10.1016/j.cmet.2020.01.002.IF=22.415(2019)
Cell Metab. 2020 Mar 3;31(3):534-548.e5. doi: 10.1016/j.cmet.2020.01.002.IF=22.415(2019) Mol Cell. 2017 Nov 16;68(4):673-685.e6. doi: 10.1016/j.molcel.2017.10.022.IF=14.548(2019)
Mol Cell. 2017 Nov 16;68(4):673-685.e6. doi: 10.1016/j.molcel.2017.10.022.IF=14.548(2019)