| Animal Research: |
| Gan,1979 Apr;70(2):209-14. | | Effect of Coadministration of Uracil or Cytosine on the Anti-Tumor Activity of Clinical Doses of 1-(2-tetrahydrofuryl)-5-fluorouracil and Level of 5-fluorouracil in Rodents.[Reference: WebLink] |
METHODS AND RESULTS:
Concentration of 5-fluorouracil (5-FU) in the tumor, blood, and various organs of AH130-bearing rats after administration of clinical doses of 1-(2-tetrahydrofuryl)-5-fluorouracil (FT-207) and uracil was examined. The concentration of 5-FU in blood was less than 0.02 microgram/ml with all combinations of FT-207 and uracil except high molar ratios of uracil to FT-207 (ratio, 5 and 10), whereas high concentrations of up to a maximum of 0.200 microgram/g on administration of uracil plus 5 or 7.5 mg/kg of FT-207 (ratio, 4), was found in the tumor. On oral administration of FT-207 plus uracil in various combinations, the highest T/B (ratio of concentration of 5-FU in the tumor to that in blood) value was obtained at a ratio of uracil to FT-207 of 4. With this combination, 5-FU concentration in the tumor, muscle, and spleen was higher than that after administration of FT-207 alone (5 mg/kg).
CONCLUSIONS:
These results suggest that at the clinical doses the optimum molar ratio of uracil to FT-207 is 4. Coadministration of Cytosine enhanced the antitumor activity of FT-207 on sarcoma-180 in mice. However, Cytosine enhanced the antitumor activity of FT-207 less than uracil and its coadministration resulted in a lower concentration of 5-FU in the tumor than coadministration of uracil. |
|
| Structure Identification: |
| Proceedings of the National Academy of ences of the United States of America, 1988, 85(12):4397-4401. | | Reactivity of cytosine and thymine in single-base-pair mismatches with hydroxylamine and osmium tetroxide and its application to the study of mutations.[Reference: WebLink] | The chemical reactivity of thymine (T), when mismatched with the bases Cytosine, guanine, and thymine, and of Cytosine (C), when mismatched with thymine, adenine, and Cytosine, has been examined.
METHODS AND RESULTS:
Heteroduplex DNAs containing such mismatched base pairs were first incubated with osmium tetroxide (for T and C mismatches) or hydroxylamine (for C mismatches) and then incubated with piperidine to cleave the DNA at the modified mismatched base. This cleavage was studied with an internally labeled strand containing the mismatched T or C, such that DNA cleavage and thus reactivity could be detected by gel electrophoresis. Cleavage at a total of 13 T and 21 C mismatches isolated (by at least three properly paired bases on both sides) single-base-pair mismatches was identified. All T or C mismatches studied were cleaved. By using end-labeled DNA probes containing T or C single-base-pair mismatches and conditions for limited cleavage, we were able to show that cleavage was at the base predicted by sequence analysis and that mismatches in a length of DNA could be readily detected by such an approach.
CONCLUSIONS:
This procedure may enable detection of all single-base-pair mismatches by use of sense and antisense probes and thus may be used to identify the mutated base and its position in a heteroduplex. | | PLoS Biol,2006 Jun;4(6):e180. | | High Guanine and Cytosine Content Increases mRNA Levels in Mammalian Cells.[Reference: WebLink] | High guanine and Cytosine content increases mRNA levels in mammalian cells. Mammalian genes are highly heterogeneous with respect to their nucleotide composition, but the functional consequences of this heterogeneity are not clear. In the previous studies, weak positive or negative correlations have been found between the silent-site guanine and Cytosine (GC) content and expression of mammalian genes. However, previous studies disregarded differences in the genomic context of genes, which could potentially obscure any correlation between GC content and expression.
METHODS AND RESULTS:
In the present work, we directly compared the expression of GC-rich and GC-poor genes placed in the context of identical promoters and UTR sequences. We performed transient and stable transfections of mammalian cells with GC-rich and GC-poor versions of Hsp70, green fluorescent protein, and IL2 genes. The GC-rich genes were expressed several-fold to over a 100-fold more efficiently than their GC-poor counterparts. This effect was not due to different translation rates of GC-rich and GC-poor mRNA. On the contrary, the efficient expression of GC-rich genes resulted from their increased steady-state mRNA levels. mRNA degradation rates were not correlated with GC content, suggesting that efficient transcription or mRNA processing is responsible for the high expression of GC-rich genes.
CONCLUSIONS:
We conclude that silent-site GC content correlates with gene expression efficiency in mammalian cells. |
|




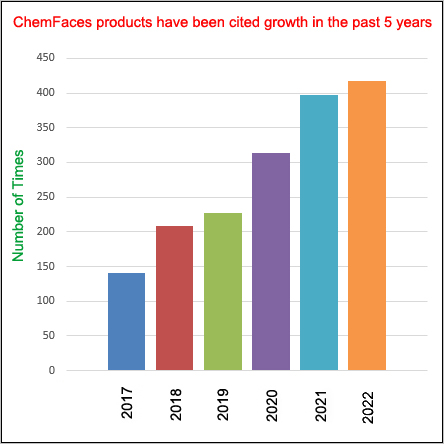
 Cell. 2018 Jan 11;172(1-2):249-261.e12. doi: 10.1016/j.cell.2017.12.019.IF=36.216(2019)
Cell. 2018 Jan 11;172(1-2):249-261.e12. doi: 10.1016/j.cell.2017.12.019.IF=36.216(2019) Cell Metab. 2020 Mar 3;31(3):534-548.e5. doi: 10.1016/j.cmet.2020.01.002.IF=22.415(2019)
Cell Metab. 2020 Mar 3;31(3):534-548.e5. doi: 10.1016/j.cmet.2020.01.002.IF=22.415(2019) Mol Cell. 2017 Nov 16;68(4):673-685.e6. doi: 10.1016/j.molcel.2017.10.022.IF=14.548(2019)
Mol Cell. 2017 Nov 16;68(4):673-685.e6. doi: 10.1016/j.molcel.2017.10.022.IF=14.548(2019)