| Abstract
Ethnopharmacological relevance: Ganoderma lucidum (GL) is an oriental medical fungus, which was used to prevent and treat many diseases. Previously, the effective compounds of Ganoderma lucidum extract (GLE) were extracted from two kinds of GL, [Ganoderma lucidum (Leyss. Ex Fr.) Karst.] and [Ganoderma sinense Zhao, Xu et Zhang], which have been used for adjuvant anti-cancer clinical therapy for more than 20 years. However, its concrete active compounds and its regulation mechanisms on tumor are unclear.
Aim of the study: In this study, we aimed to identify the main active compounds from GLE and to investigate its anti-cancer mechanisms via drug-target biological network construction and prediction.
Materials and methods: The main active compounds of GLE were identified by HPLC, EI-MS and NMR, and the compounds related targets were predicted using docking program. To investigate the functions of GL holistically, the active compounds of GL and related targets were predicted based on four public databases. Subsequently, the Identified-Compound-Target network and Predicted-Compound-Target network were constructed respectively, and they were overlapped to detect the hub potential targets in both networks. Furthermore, the qRT-PCR and western-blot assays were used to validate the expression levels of target genes in GLE treated Hepa1-6-bearing C57 BL/6 mice.
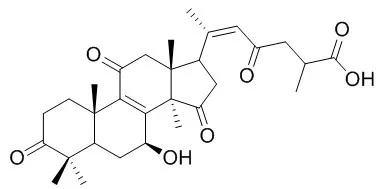
Results: In our work, 12 active compounds of GLE were identified, including Ganoderic acid A, Ganoderenic acid A, Ganoderic acid B, Ganoderic acid H, Ganoderic acid C2, Ganoderenic acid D, Ganoderic acid D, Ganoderenic acid G, Ganoderic acid Y, Kaemferol, Genistein and Ergosterol. Using the docking program, 20 targets were mapped to 12 compounds of GLE. Furthermore, 122 effective active compounds of GL and 116 targets were holistically predicted using public databases. Compare with the Identified-Compound-Target network and Predicted-Compound-Target network, 6 hub targets were screened, including AR, CHRM2, ESR1, NR3C1, NR3C2 and PGR, which was considered as potential markers and might play important roles in the process of GLE treatment. GLE effectively inhibited tumor growth in Hepa1-6-bearing C57 BL/6 mice. Finally, consistent with the results of qRT-PCR data, the results of western-blot assay demonstrated the expression levels of PGR and ESR1 were up-regulated, as well as the expression levels of NR3C2 and AR were down-regulated, while the change of NR3C1 and CHRM2 had no statistical significance.
Conclusions: The results indicated that these 4 hub target genes, including NR3C2, AR, ESR1 and PGR, might act as potential markers to evaluate the curative effect of GLE treatment in tumor. And, the combined data provide preliminary study of the pharmacological mechanisms of GLE, which may be a promising potential therapeutic and chemopreventative candidate for anti-cancer.
Keywords: Cancer; Ergosterol (PubChem CID: 444679); Ganoderenic acid A (PubChem CID: 6442088); Ganoderenic acid D (PubChem CID: 101600080); Ganoderenic acid G (PubChem CID: 14193981); Ganoderic acid A (PubChem CID: 471002); Ganoderic acid B (PubChem CID: 471003); Ganoderic acid C2 (PubChem CID: 57396771); Ganoderic acid D (PubChem CID: 102004379); Ganoderic acid H (PubChem CID: 471005); Ganoderic acid Y (PubChem CID: 57397445); Ganoderma lucidum; Genistein (PubChem CID: 5281377); Kaemferol (PubChem CID: 5280863); Network pharmacology; Target gene. |



 Cell. 2018 Jan 11;172(1-2):249-261.e12. doi: 10.1016/j.cell.2017.12.019.IF=36.216(2019)
Cell. 2018 Jan 11;172(1-2):249-261.e12. doi: 10.1016/j.cell.2017.12.019.IF=36.216(2019) Cell Metab. 2020 Mar 3;31(3):534-548.e5. doi: 10.1016/j.cmet.2020.01.002.IF=22.415(2019)
Cell Metab. 2020 Mar 3;31(3):534-548.e5. doi: 10.1016/j.cmet.2020.01.002.IF=22.415(2019) Mol Cell. 2017 Nov 16;68(4):673-685.e6. doi: 10.1016/j.molcel.2017.10.022.IF=14.548(2019)
Mol Cell. 2017 Nov 16;68(4):673-685.e6. doi: 10.1016/j.molcel.2017.10.022.IF=14.548(2019)