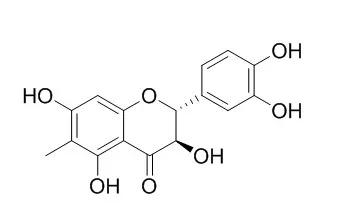
| Structure Identification: |
| Adv Bioinformatics. 2014;2014:903246. | | Pharmacophore Modeling and Molecular Docking Studies on Pinus roxburghii as a Target for Diabetes Mellitus.[Pubmed: 25114678] | The present study attempts to establish a relationship between ethnopharmacological claims and bioactive constituents present in Pinus roxburghii against all possible targets for diabetes through molecular docking and to develop a pharmacophore model for the active target.
METHODS AND RESULTS:
The process of molecular docking involves study of different bonding modes of one ligand with active cavities of target receptors protein tyrosine phosphatase 1-beta (PTP-1β), dipeptidyl peptidase-IV (DPP-IV), aldose reductase (AR), and insulin receptor (IR) with help of docking software Molegro virtual docker (MVD). From the results of docking score values on different receptors for antidiabetic activity, it is observed that constituents, namely, secoisoresinol, pinoresinol, and Cedeodarin, showed the best docking results on almost all the receptors, while the most significant results were observed on AR. Then, LigandScout was applied to develop a pharmacophore model for active target. LigandScout revealed that 2 hydrogen bond donors pointing towards Tyr 48 and His 110 are a major requirement of the pharmacophore generated.
CONCLUSIONS:
In our molecular docking studies, the active constituent, secoisoresinol, has also shown hydrogen bonding with His 110 residue which is a part of the pharmacophore. The docking results have given better insights into the development of better aldose reductase inhibitor so as to treat diabetes related secondary complications. | | Planta Med. 1981 Sep;43(1):82-5. | | Dihydroflavanonols from Cedrus deodara, A (13)C NMR study.[Pubmed: 17402014] |
METHODS AND RESULTS:
High resolution (13)C NMR study of taxifolin, Cedeodarin, cedrin and their methyl ethers allowed unambiguous placement of the Me in 5,7-dihydroxyflavanonol nucleus, besides providing other valuable information on the substitution pattern in the molecule. |
|



 Cell. 2018 Jan 11;172(1-2):249-261.e12. doi: 10.1016/j.cell.2017.12.019.IF=36.216(2019)
Cell. 2018 Jan 11;172(1-2):249-261.e12. doi: 10.1016/j.cell.2017.12.019.IF=36.216(2019) Cell Metab. 2020 Mar 3;31(3):534-548.e5. doi: 10.1016/j.cmet.2020.01.002.IF=22.415(2019)
Cell Metab. 2020 Mar 3;31(3):534-548.e5. doi: 10.1016/j.cmet.2020.01.002.IF=22.415(2019) Mol Cell. 2017 Nov 16;68(4):673-685.e6. doi: 10.1016/j.molcel.2017.10.022.IF=14.548(2019)
Mol Cell. 2017 Nov 16;68(4):673-685.e6. doi: 10.1016/j.molcel.2017.10.022.IF=14.548(2019)