| Structure Identification: |
| J Biochem. 2002 Nov;132(5):819-28. | | Three-dimensional structural model analysis of the binding site of an inhibitor, nervonic acid, of both DNA polymerase beta and HIV-1 reverse transcriptase.[Pubmed: 12417034] |
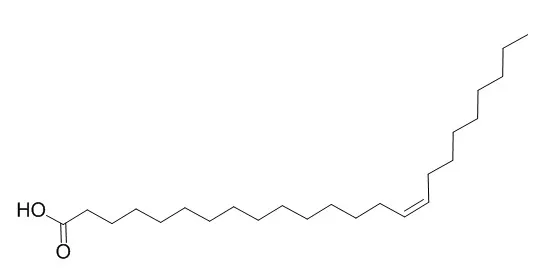
Previously, we reported the three-dimensional molecular interactions of Nervonic acid (NA) with mammalian DNA polymerase beta (pol beta) [Mizushina et al. (1998) J. Biol. Chem. 274, 25599-25607].
METHODS AND RESULTS:
By three-dimensional structural model analysis and comparison with the spatial positioning of specific amino acids binding to NA on pol beta (Leu11, Lys35, His51, and Thr79), we obtained supplementary information that allowed us to build a structural model of human immunodeficiency virus type-1 reverse transcriptase (HIV-1 RT). In HIV-1 RT, Leu100, Lys65, His235, and Thr386 corresponded to these four amino acid residues.
CONCLUSIONS:
These results suggested that the NA binding domains of pol beta and HIV-1 RT are three-dimensionally very similar. The effects of NA on HIV-1 RT are thought to be same as those on pol beta in binding to the rhombus of the four amino acid residues. NA dose-dependently inhibited the HIV-1 RT activity. For binding to pol beta, the kinetics were competitive when the rhombus was present on the DNA binding site. However, as the rhombus in HIV-1 RT was not present in the DNA binding site, the three-dimensional structure of the DNA binding site must be distorted, and subsequently the enzyme is inhibited non-competitively. |
|



 Cell. 2018 Jan 11;172(1-2):249-261.e12. doi: 10.1016/j.cell.2017.12.019.IF=36.216(2019)
Cell. 2018 Jan 11;172(1-2):249-261.e12. doi: 10.1016/j.cell.2017.12.019.IF=36.216(2019) Cell Metab. 2020 Mar 3;31(3):534-548.e5. doi: 10.1016/j.cmet.2020.01.002.IF=22.415(2019)
Cell Metab. 2020 Mar 3;31(3):534-548.e5. doi: 10.1016/j.cmet.2020.01.002.IF=22.415(2019) Mol Cell. 2017 Nov 16;68(4):673-685.e6. doi: 10.1016/j.molcel.2017.10.022.IF=14.548(2019)
Mol Cell. 2017 Nov 16;68(4):673-685.e6. doi: 10.1016/j.molcel.2017.10.022.IF=14.548(2019)