| Kinase Assay: |
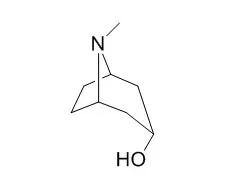
| PLoS One. 2013 Sep 25;8(9):e74777. | | Tropine forming tropinone reductase gene from Withania somnifera (Ashwagandha): biochemical characteristics of the recombinant enzyme and novel physiological overtones of tissue-wide gene expression patterns.[Pubmed: 24086372] | Withania somnifera is one of the most reputed medicinal plants of Indian systems of medicine synthesizing diverse types of secondary metabolites such as withanolides, alkaloids, withanamides etc. Present study comprises cloning and E. coli over-expression of a tropinone reductase gene (WsTR-I) from W. somnifera, and elucidation of biochemical characteristics and physiological role of tropinone reductase enzyme in tropane alkaloid biosynthesis in aerial tissues of the plant.
METHODS AND RESULTS:
The recombinant enzyme was demonstrated to catalyze NADPH-dependent tropinone to Tropine conversion step in tropane metabolism, through TLC, GC and GC-MS-MS analyses of the reaction product. The functionally active homodimeric ~60 kDa enzyme catalyzed the reaction in reversible manner at optimum pH 6.7. Catalytic kinetics of the enzyme favoured its forward reaction (Tropine formation). Comparative 3-D models of landscape of the enzyme active site contours and tropinone binding site were also developed. Tissue-wide and ontogenic stage-wise assessment of WsTR-I transcript levels revealed constitutive expression of the gene with relatively lower abundance in berries and young leaves. The tissue profiles of WsTR-I expression matched those of Tropine levels. The data suggest that, in W. somnifera, aerial tissues as well possess tropane alkaloid biosynthetic competence. In vivo feeding of U-[(14)C]-sucrose to orphan shoot (twigs) and [(14)C]-chasing revealed substantial radiolabel incorporation in tropinone and Tropine, confirming the de novo synthesizing ability of the aerial tissues.
CONCLUSIONS:
This inherent independent ability heralds a conceptual novelty in the backdrop of classical view that these tissues acquire the alkaloids through transportation from roots rather than synthesis. The TR-I gene expression was found to be up-regulated on exposure to signal molecules (methyl jasmonate and salicylic acid) and on mechanical injury. The enzyme's catalytic and structural properties as well as gene expression profiles are discussed with respect to their physiological overtones. |
|
| Structure Identification: |
| Biochem J. 1995 Apr 15;307 ( Pt 2):603-8. | | Tropine dehydrogenase: purification, some properties and an evaluation of its role in the bacterial metabolism of tropine.[Pubmed: 7733902] | Tropine dehydrogenase was induced by growth of Pseudomonas AT3 on aTropine, Tropine or tropinone. It was NADP(+)-dependent and gave no activity with NAD+. The enzyme was very unstable but a rapid purification procedure using affinity chromatography that gave highly purified enzyme was developed.
METHODS AND RESULTS:
The enzyme gave a single band on isoelectric focusing with an isoelectric point at approximately pH 4. The native enzyme had an M(r) of 58,000 by gel filtration and 28,000 by SDS/PAGE and therefore consists of two subunits of equal size. The enzyme displayed a narrow range of specificity and was active with Tropine and norTropine but not with pseudoTropine, pseudonorTropine, or a number of related compounds. The apparent Kms were 6.06 microM for Tropine and 73.4 microM for norTropine with the specificity constant (Vmax/Km) for Tropine 7.8 times that for pseudoTropine. The apparent Km for NADP+ was 48 microM.
CONCLUSIONS:
The deuterium of [3-2H]Tropine and [3-2H]pseudoTropine was retained when these compounds were converted into 6-hydroxycyclohepta-1,4-dione, an intermediate in Tropine catabolism, showing that the Tropine dehydrogenase, although induced by growth on Tropine, is not involved in the catabolic pathway for this compound. 6-Hydroxycyclohepta-1,4-dione was also implicated as an intermediate in the pathways for pseudoTropine and tropinone catabolism. |
|



 Cell. 2018 Jan 11;172(1-2):249-261.e12. doi: 10.1016/j.cell.2017.12.019.IF=36.216(2019)
Cell. 2018 Jan 11;172(1-2):249-261.e12. doi: 10.1016/j.cell.2017.12.019.IF=36.216(2019) Cell Metab. 2020 Mar 3;31(3):534-548.e5. doi: 10.1016/j.cmet.2020.01.002.IF=22.415(2019)
Cell Metab. 2020 Mar 3;31(3):534-548.e5. doi: 10.1016/j.cmet.2020.01.002.IF=22.415(2019) Mol Cell. 2017 Nov 16;68(4):673-685.e6. doi: 10.1016/j.molcel.2017.10.022.IF=14.548(2019)
Mol Cell. 2017 Nov 16;68(4):673-685.e6. doi: 10.1016/j.molcel.2017.10.022.IF=14.548(2019)